SATAY Experimental Process
SATAY Experimental Process¶
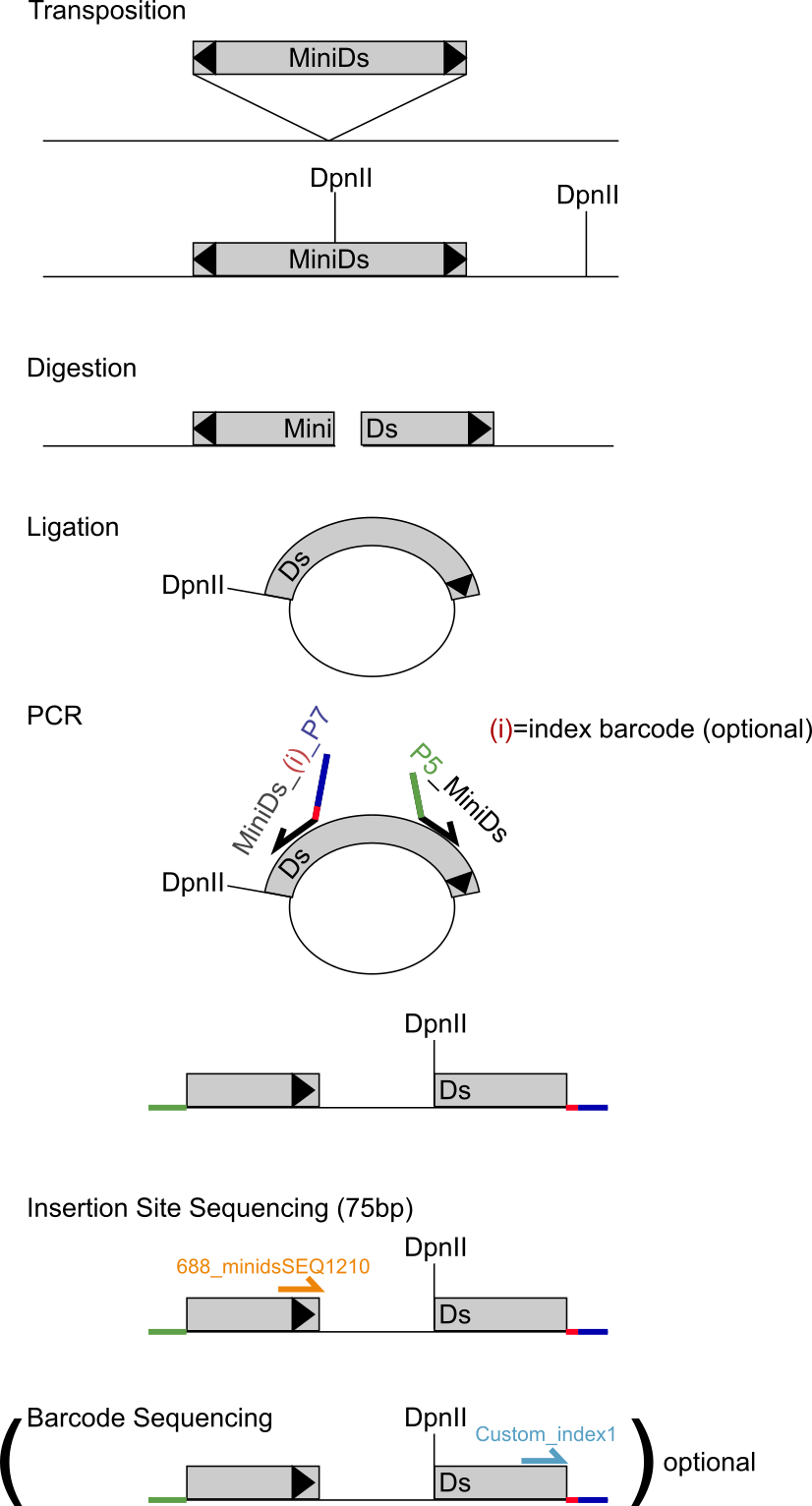
The process of SATAY starts with inserting a plasmid in the cells that contains a transposase (TPase) and the transposon (MiniDs) flanked on both sides by adenine (ADE). The transposon has a specific, known, sequence that codes for the transposase that cuts the transposon from the plasmid (or DNA) to (another part of) the DNA.
Simplified example for the transposon insertion plasmid.
The MiniDs transposon is cut loose from the plasmid and randomly inserted in the DNA of the host cell.
If the transposon is inserted in a gene, the gene can still be transcribed by the ribosomes, but typically cannot be (properly) translated in a functional protein.
The genomic DNA (with the transposon) is cut in pieces for sequencing using enzymes, for example DpnII.
This cuts the DNA in many small pieces (e.g. each 75bp long) and it always cuts the transposon in two parts (i.e. digestion of the DNA).
Each of the two halves of the cut transposon, together with the part of the gene where the transposon is inserted in, is ligated meaning that it is folded in a circle.
A part of the circle is then the half transposon and the rest of the circle is a part of the gene where the transposon is inserted in.
Using PCR and primers, this can then be unfolded by cutting the circle at the halved transposon.
The part of the gene is then between the transposon quarters.
Since the sequence of the transposon is known, the part of the gene can be extracted.
This is repeated for the other half of the transposon that includes the other part of the gene.
When both parts of the gene are known, the sequence from the original gene can be determined.